This is based on papers of Tchetgen Tchetgen and co-authors, mostly based on Xu Shi’s talk on proximal causal inference.
Unobserved confounder
Unobserved confounder is the problem for observational studies. Assuming controlling for all relevant confounders (), we have an unobserved confounder that affects both treatment and outcome . The causal diagram is as follows:
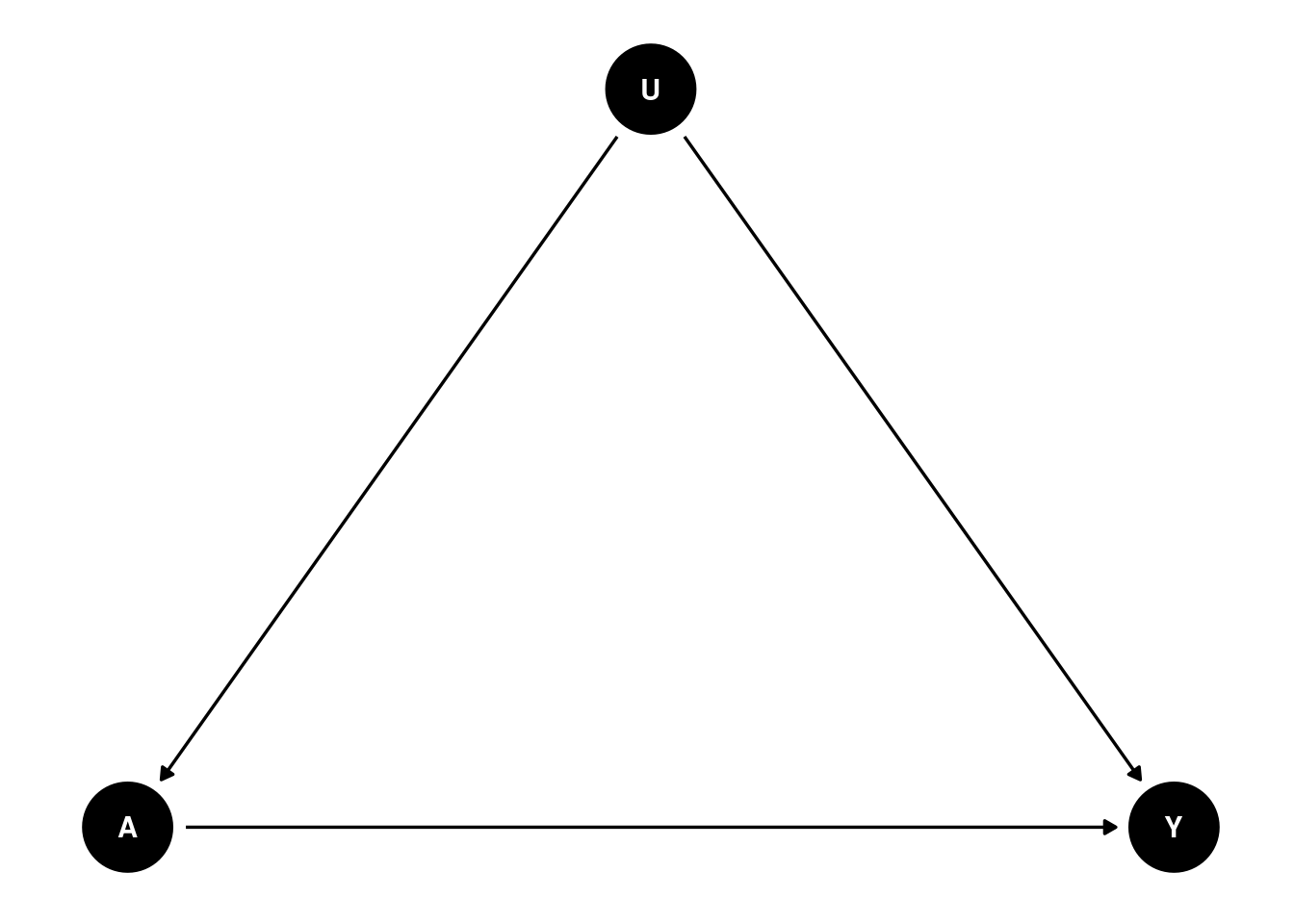
In this case, we have biased estimates of the treatment effect, regressing on controlling for .
Proximal causal inference
Proximal causal inference (also called negative controls) is a framework that allows us to estimate the causal effect of treatment on outcome while accounting for unobserved confounders.
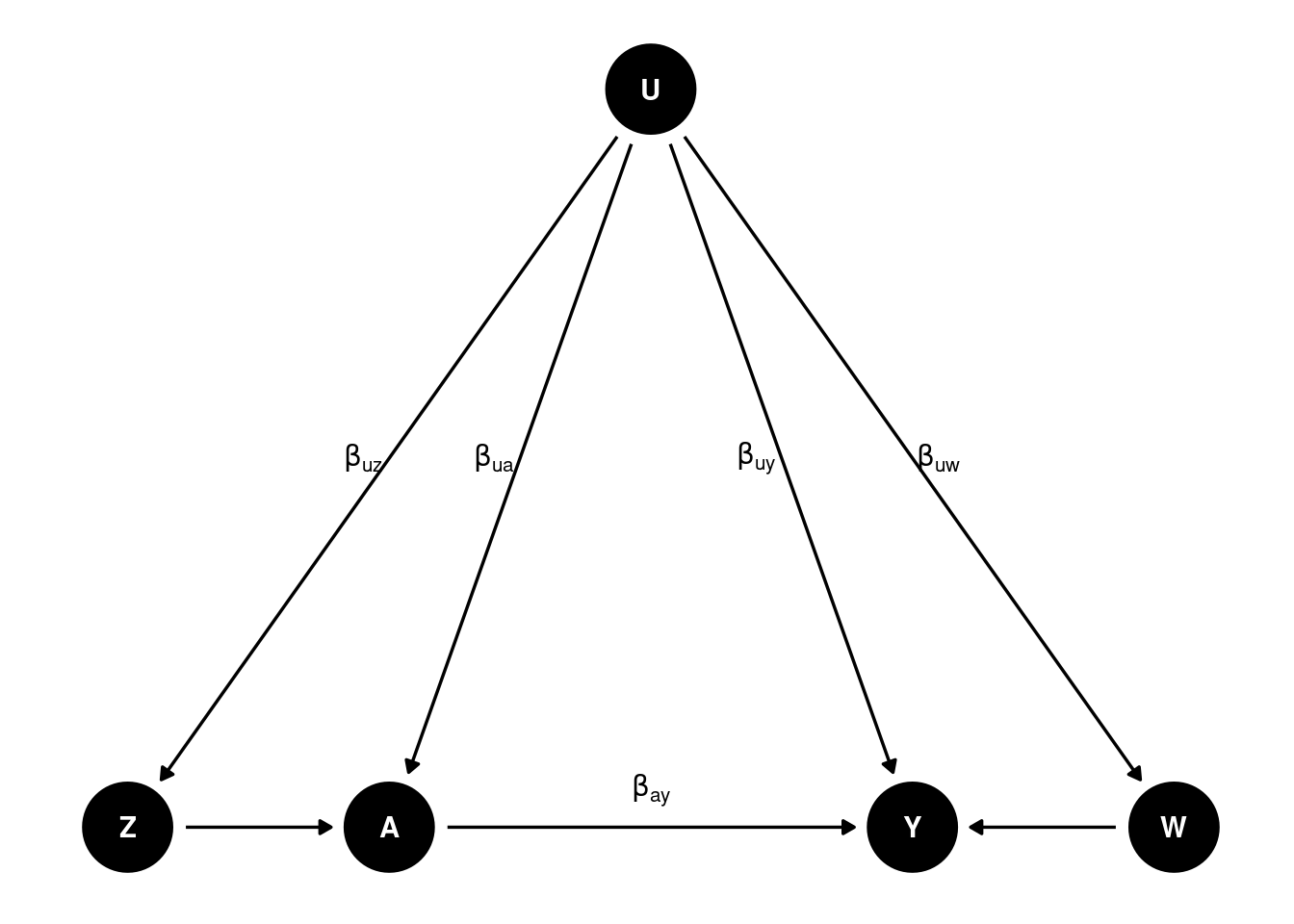
We’ll need a DAG like this:
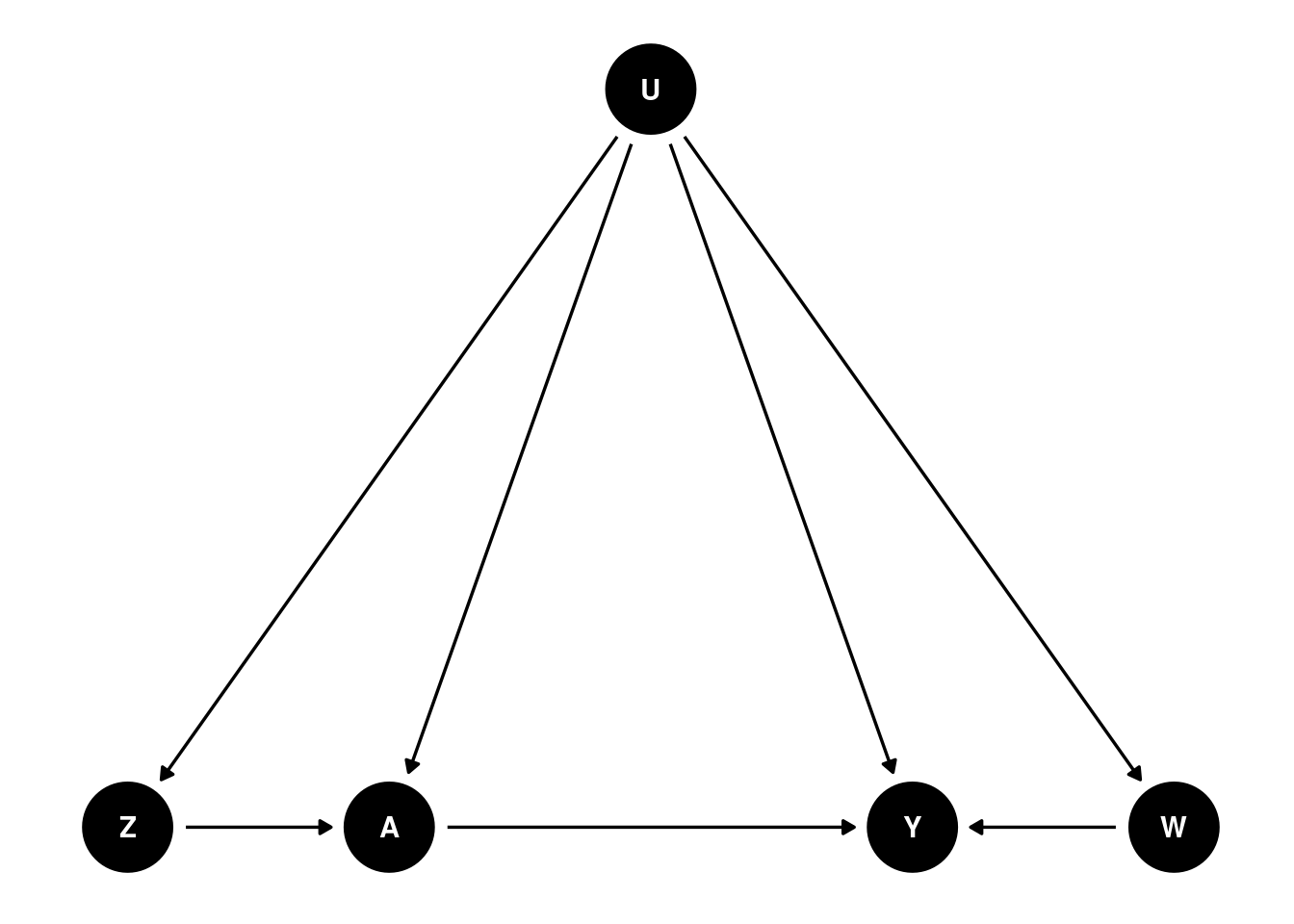
Basically we need two proxies for to control for confounding. is called Negative Control for Exposure (NCE) and is called Negative Control for Outcome (NCO). There is no link form to , and no link from to or .
is an NCE if and , meaning that satisfies the exclusion restriction that there is no direct link from to , and that is independent of given .
is an NCO if and , meaning that and have no direct link to .
Why this works

If we regress on , then we get , which is biased. How do we recover ?
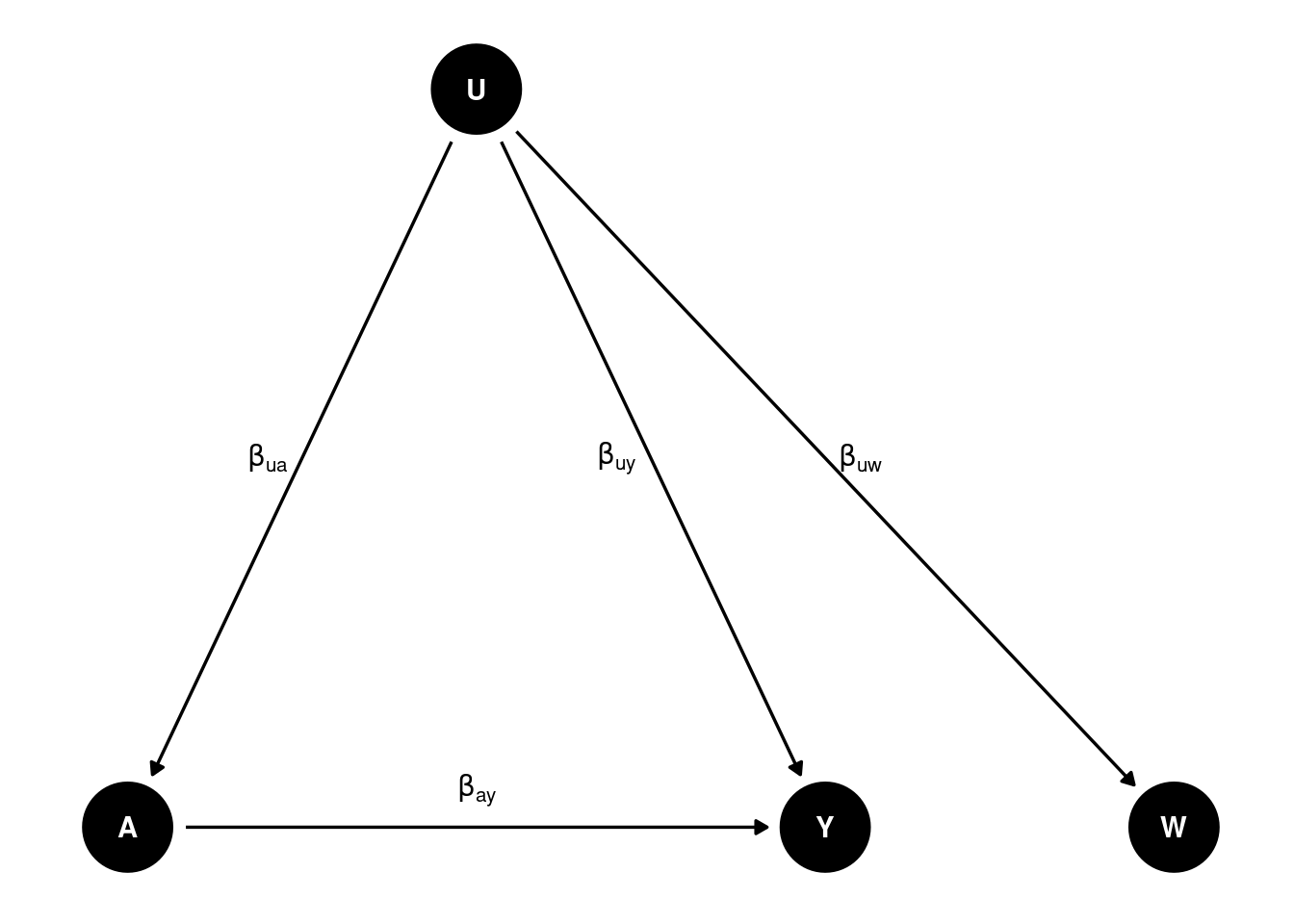
Suppose we have as proxy of and , then we can get the true effect. We can do regression of on , and then on . The first regression gives us , and the second regression gives us . We can then get from the difference of the two coefficients. This is similar to a DiD setting. Say is , and assume the effect of is the same on and , then effect of on by the difference of regressing on and on .
However, most likely . Then we need another proxy of to recover the true effect. This is also called “double proxy”, or “double negative control”.
A linear model
Suppose we have the following linear model:
is the true effect of on .
From the last two regressions we can recover :
If we define (regression coefficient of on the regression) and , (regression coefficient of on the regression), similarly for and , then we have
Or, we can look at this as a two-stage least squares (2SLS) regression.
We can replace with the predicted value of from the second equation:
We first regress on and , and then regress on and the predicted value of .
A nonparametric model
We don’t have to have a linear model, this can be done with a nonparametric model as well.
where .
That is, we can estimate as a function of and , and nonparametrically. This function is called outcome bridge function. In the linear case, it is .
How to find proximal variables
Tchetgen Tchetgen and co-authors come up with a Data-driven Automated Negative Control Estimation (DANCE) algorithm to find negative controls. The idea is to use a machine learning algorithm to find the variables that are correlated with the treatment and the outcome, but not with the treatment and the outcome together. This is similar to the idea of finding instrumental variables.
example
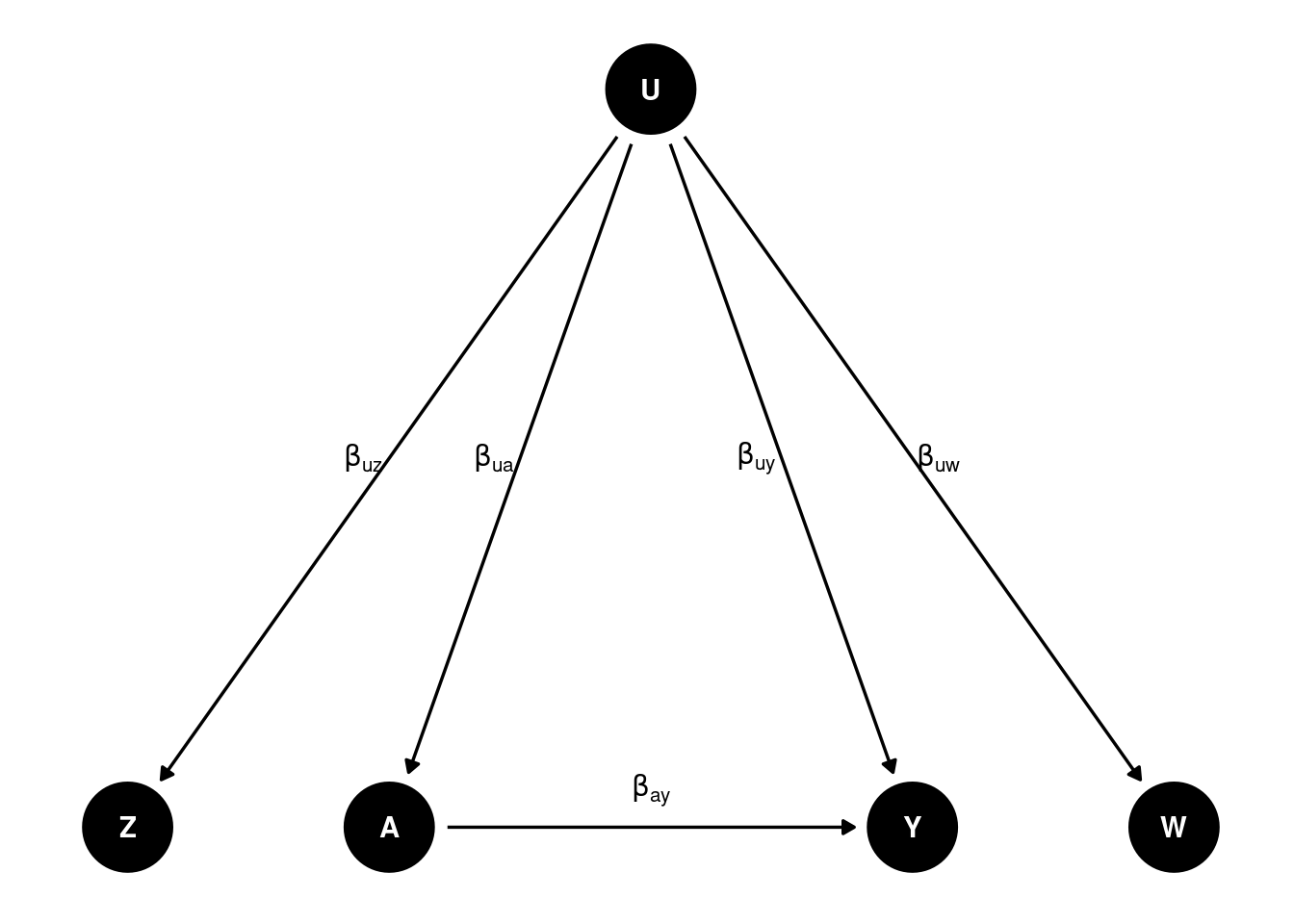
Let’s generated data based on this DAG:
set.seed(123)
n <- 5000
# Set coefficients
b_uw <- 0.5 # effect of U on W
b_uy <- 0.5 # effect of U on Y
b_ay <- 0.5 # effect of A on Y
b_uz <- 0.5 # effect of U on Z
b_ua <- 0.5 # effect of U on A
b_za <- 0.5 # effect of Z on A
U <- rnorm(n)
W <- b_uw * U + rnorm(n) # W is a function of U
Z <- b_uz * U + rnorm(n) # Z is a function of U
A <- b_ua * U + b_za * Z + rnorm(n) # A is a function of U and Z
Y <- b_uy * U + b_ay * A + rnorm(n) # Y is a function of U and A
# A <- rbinom(n, 1, plogis(-1 + U / 2)) # A is a binary treatment
# Create a data frame with the generated variables
data <- data.frame(W = W, Z = Z, A = A, Y = Y)If we run regression of on , , ,
m1 <- lm(Y ~ A + Z + W, data = data)
summary(m1)##
## Call:
## lm(formula = Y ~ A + Z + W, data = data)
##
## Residuals:
## Min 1Q Median 3Q Max
## -4.5836 -0.7167 0.0005 0.7305 4.4866
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -0.006347 0.015279 -0.415 0.678
## A 0.660021 0.014123 46.734 < 2e-16 ***
## Z 0.085004 0.016650 5.105 3.43e-07 ***
## W 0.132798 0.014165 9.375 < 2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 1.08 on 4996 degrees of freedom
## Multiple R-squared: 0.4556, Adjusted R-squared: 0.4553
## F-statistic: 1394 on 3 and 4996 DF, p-value: < 2.2e-16We don’t get the true effect of on .
If we do the 2sls using and , then we get the true effect.
# Linear (Y, W) Case
# Perform the first-stage linear regression W ~ A + Z
m1 <- lm(W ~ A + Z, data = data)
# Compute the estimated E[W|A,Z]
S <- m1$fitted.values
# Perform the second-stage linear regression Y ~ A + E[W|A,Z]
m2 <- lm(Y ~ A + S, data = data)
summary(m2)##
## Call:
## lm(formula = Y ~ A + S, data = data)
##
## Residuals:
## Min 1Q Median 3Q Max
## -4.5477 -0.7309 0.0059 0.7395 4.3715
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -0.005049 0.015408 -0.328 0.743
## A 0.489342 0.043210 11.325 < 2e-16 ***
## S 1.143480 0.199171 5.741 9.96e-09 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 1.089 on 4997 degrees of freedom
## Multiple R-squared: 0.446, Adjusted R-squared: 0.4458
## F-statistic: 2012 on 2 and 4997 DF, p-value: < 2.2e-16